Second in popularity for the expansion of haplogroup N1a-L392 (ca. 4400 BC) is, apparently, the association with Turkic, and by extension with Micro-Altaic, after the Uralic link preferred in Europe; at least among certain eastern researchers.
New paper in a recently created journal, by the same main author of the group proposing that Scythians of hg. N1c were Turkic speakers: On the origins of the Sakhas’ paternal lineages: Reconciliation of population genetic / ancient DNA data, archaeological findings and historical narratives, by Tikhonov, Gurkan, Demirdov, and Beyoglu, Siberian Research (2019).
Interesting excerpts:
According to the views of a number of authoritative researchers, the Yakut ethnos was formed in the territory of Yakutia as a result of the mixing of people from the south and the autochthonous population [34].
These three major Sakha paternal lineages may have also arrived in Yakutia at different times and/ or from different places and/or with a difference in several generations instead, or perhaps Y-chromosomal STR mutations may have taken place in situ in Yakutia. Nevertheless, the immediate common ancestor(s) from the Asian Steppe of these three most prevalent Sakha Y-chromosomal STR haplotypes possibly lived during the prominence of the Turkic Khaganates, hence the near-perfect matches observed across a wide range of Eurasian geography, including as far as from Cyprus in the West to Liaoning, China in the East, then Middle Lena in the North and Afghanistan in the South (Table 3 and Figure 5). There may also be haplotypes closely-related to ‘the dominant Elley line’ among Karakalpaks, Uzbeks and Tajiks, however, limitations in the loci coverage for the available dataset (only eight Y-chromosomal STR loci) precludes further conclusions on this matter [25].
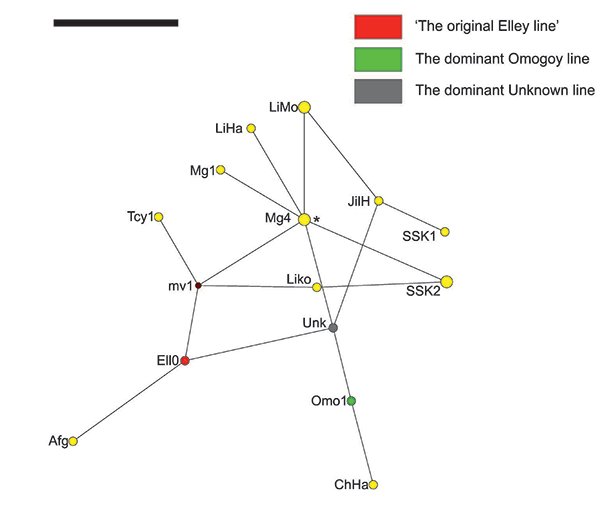
According to the results presented here, very similar Y-STR haplotypes to that of the original Elley line were found in the west: Afghanistan and northern Cyprus, and in the east: Liaoning Province, China and Ulaanbaator, Northern Mongolia. In the case of the dominant Omogoy line, very closely matching haplotypes differing by a single mutational step were found in the city of Chifen of the Jirin Province, China. The widest range of similar haplotypes was found for the Yakut haplotype Unknown: In Mongolia, China and South Korea. For instance, haplotypes differing by a single step mutation were found in Northern Mongolia (Khalk, Darhad, Uryankhai populations), Ulaanbaator (Khalk) and in the province of Jirin, China (Han population).

(Vil), Omogoy (Omo), Eurasian (Eur) and Xiongnu (Xuo) Y-chromosomal STR haplotypes and that for a representative ancient DNA sample (Ch0 or DSQ04) from the Upper Xiajiadian Culture
recovered from the Inner Mongolia Autonomous Region, China.
Notably, Tat-C-bearing Y-chromosomes were also observed in ancient DNA samples from the 2700-3000 years-old Upper Xiajiadian culture in Inner Mongolia, as well as those from the Serteya II site at the Upper Dvina region in Russia and the ‘Devichyi gory’ culture of long barrow burials at the Nevel’sky district of Pskovsky region in Russia. A 14-loci Y-chromosomal STR median-joining network of the most prevalent Sakha haplotypes and a Tat-C-bearing haplotype from one of the ancient DNA samples recovered from the Upper Xiajiadian culture in Inner Mongolia (DSQ04) revealed that the contemporary Sakha haplotype ‘Xuo’ (Table 2, Haplotype ID “Xuo”) classified as that of ‘the Xiongnu clan’ in our current study, was the closest to the ancient Xiongnu haplotype (Figure 6). TMRCA estimate for this 14-loci Y-chromosomal STR network was 4357 ± 1038 years or 2341 ± 1038 BCE, which correlated well with the Upper Xiajiadian culture that was dated to the Late Bronze Age (700-1000 BCE).
NOTE. Also interesting from the paper seems to be the proportion of E1b1b among admixed Russian populations, in a proportion similar to R1a or I2a(xI2a1).
It is tempting to associate the prevalent presence of N1c-L392 in ancient Siberian populations with the expansion of Altaic, by simplistically linking the findings (in chronological order) near Lake Baikal (Damgaard et al. 2018), Upper Xiajiadian (Cui et al. 2013), among Khövsgöl (Jeong et al. 2018), in Huns (Damgaard et al. 2018), and in Mongolic-speaking Avars (Csáky et al. 2019).
However, its finding among Palaeo-Laplandic peoples in the Kola peninsula ca. 1500 BC (Lamnidis et al. 2018) and among Palaeo-Siberian populations near the Yana River (Sikora et al. 2018) ca. AD 1200 should be enough to accept the hypothesis of ancestral waves of expansion of the haplogroup over northern Eurasia, with acculturation and further expansions in the different regions since the Iron Age (see more on its potential expansion waves).
Also, a simple look at the TMRCA and modern distribution was enough to hypothesize long ago the lack of connection of N1c-L392 with Altaic or Uralic peoples. From Ilumäe et al. (2016):
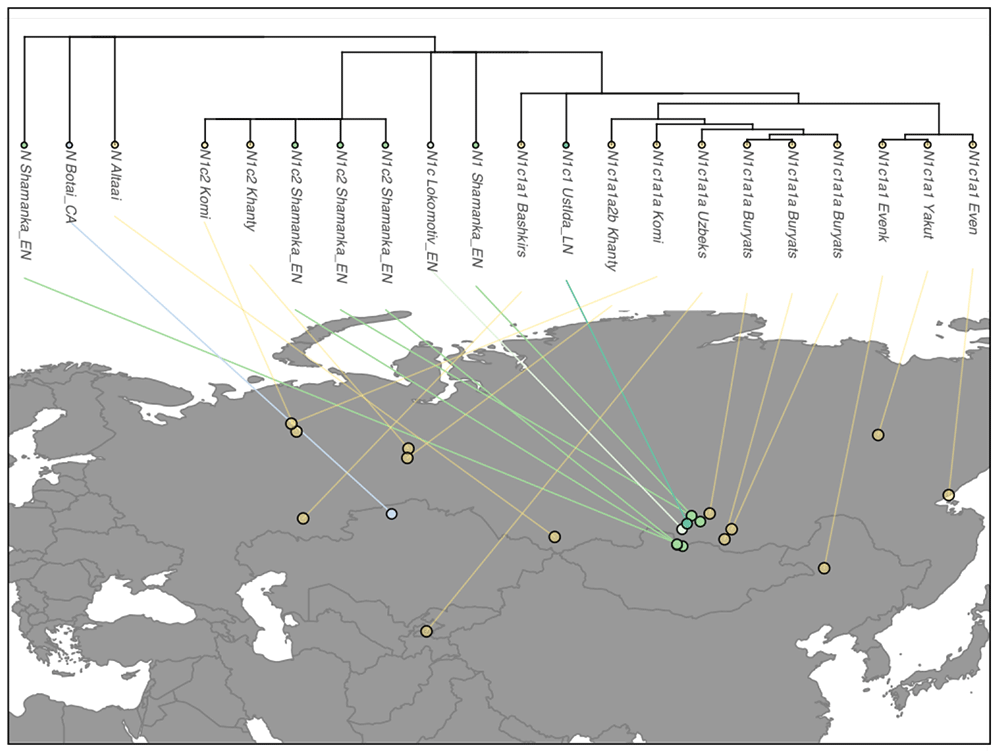
Previous research has shown that Y chromosomes of the Turkic-speaking Yakuts (Sakha) belong overwhelmingly to hg N3 (formerly N1c1). We found that nearly all of the more than 150 genotyped Yakut N3 Y chromosomes belong to the N3a2-M2118 clade, just as in the Turkic-speaking Dolgans and the linguistically distant Tungusic-speaking Evenks and Evens living in Yakutia (Table S2). Hence, the N3a2 patrilineage is a prime example of a male population of broad central Siberian ancestry that is not intrinsic to any linguistically defined group of people. Moreover, the deepest branch of hg N3a2 is represented by a Lebanese and a Chinese sample. This finding agrees with the sequence data from Hallast et al., where one Turkish Y chromosome was also assigned to the same sub-clade. Interestingly, N3a2 was also found in one Bhutan individual who represents a separate sub-lineage in the clade. These findings show that although N3a2 reflects a recent strong founder effect primarily in central Siberia (Yakutia, Sakha), the sub-clade has a much wider distribution area with incidental occurrences in the Near East and South Asia.
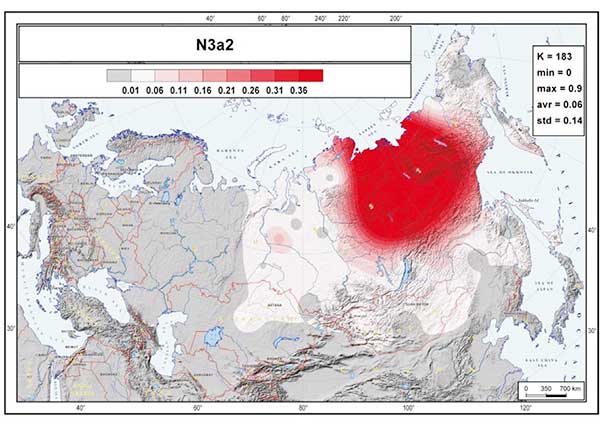
The most striking aspect of the phylogeography of hg N is the spread of the N3a3’6-CTS6967 lineages. Considering the three geographically most distant populations in our study—Chukchi, Buryats, and Lithuanians—it is remarkable to find that about half of the Y chromosome pool of each consists of hg N3 and that they share the same sub-clade N3a3’6. The fractionation of N3a3’6 into the four sub-clades that cover such an extraordinarily wide area occurred in the mid-Holocene, about 5.0 kya (95% CI = 4.4–5.7 kya). It is hard to pinpoint the precise region where the split of these lineages occurred. It could have happened somewhere in the middle of their geographic spread around the Urals or further east in West Siberia, where current regional diversity of hg N sub-lineages is the highest (Figure 1B). Yet, it is evident that the spread of the newly arisen sub-clades of N3a3’6 in opposing directions happened very quickly. Today, it unites the East Baltic, East Fennoscandia, Buryatia, Mongolia, and Chukotka-Kamchatka (Beringian) Eurasian regions, which are separated from each other by approximately 5,000–6,700 km by air. N3a3’6 has high frequencies in the patrilineal pools of populations belonging to the Altaic, Uralic, several Indo-European, and Chukotko-Kamchatkan language families. There is no generally agreed, time-resolved linguistic tree that unites these linguistic phyla. Yet, their split is almost certainly at least several millennia older than the rather recent expansion signal of the N3a3’6 sub-clade, suggesting that its spread had little to do with linguistic affinities of men carrying the N3a3’6 lineages.
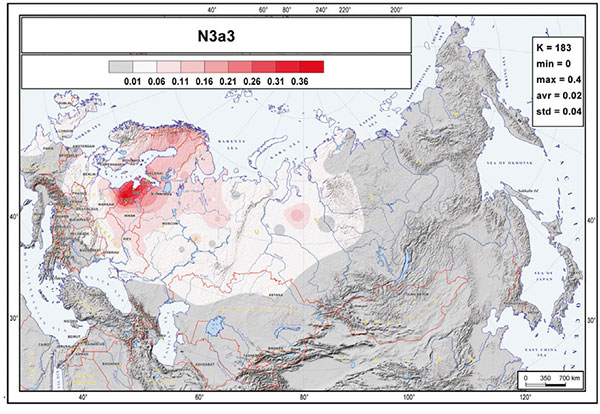
It was thus clear long ago that N1c-L392 lineages must have expanded explosively in the 5th millennium through Northern Eurasia, probably from a region to the north of Lake Baikal, and that this expansion – and succeeding ones through Northern Eurasia – may not be associated to any known language group until well into the common era.
Related
- Magyar tribes brought R1a-Z645, I2a-L621, and N1a-L392(xB197) lineages to the Carpathian Basin
- R1a-Z280 and R1a-Z93 shared by ancient Finno-Ugric populations; N1c-Tat expanded with Micro-Altaic
- Scytho-Siberians of Aldy-Bel and Sagly, of haplogroup R1a-Z93, Q1b-L54, and N
- Mitogenomes from Avar nomadic elite show Inner Asian origin
- The complex origin of Samoyedic-speaking populations
- Corded Ware—Uralic (IV): Hg R1a and N in Finno-Ugric and Samoyedic expansions
- Corded Ware—Uralic (III): “Siberian ancestry” and Ugric-Samoyedic expansions
- Corded Ware—Uralic (II): Finno-Permic and the expansion of N-L392/Siberian ancestry
- The traditional multilingualism of Siberian populations
- Iron Age bottleneck of the Proto-Fennic population in Estonia
- Y-DNA haplogroups of Tuvinian tribes show little effect of the Mongol expansion
- Corded Ware—Uralic (I): Differences and similarities with Yamna
- Haplogroup R1a and CWC ancestry predominate in Fennic, Ugric, and Samoyedic groups
- On the origin and spread of haplogroup R1a-Z645 from eastern Europe
- Oldest N1c1a1a-L392 samples and Siberian ancestry in Bronze Age Fennoscandia