New paper (behind paywall), Ancient genomes from Iceland reveal the making of a human population, by Ebenesersdóttir et al. Science (2018) 360(6392):1028-1032.
Abstract and relevant excerpts (emphasis mine):
Opportunities to directly study the founding of a human population and its subsequent evolutionary history are rare. Using genome sequence data from 27 ancient Icelanders, we demonstrate that they are a combination of Norse, Gaelic, and admixed individuals. We further show that these ancient Icelanders are markedly more similar to their source populations in Scandinavia and the British-Irish Isles than to contemporary Icelanders, who have been shaped by 1100 years of extensive genetic drift. Finally, we report evidence of unequal contributions from the ancient founders to the contemporary Icelandic gene pool. These results provide detailed insights into the making of a human population that has proven extraordinarily useful for the discovery of genotype-phenotype associations.
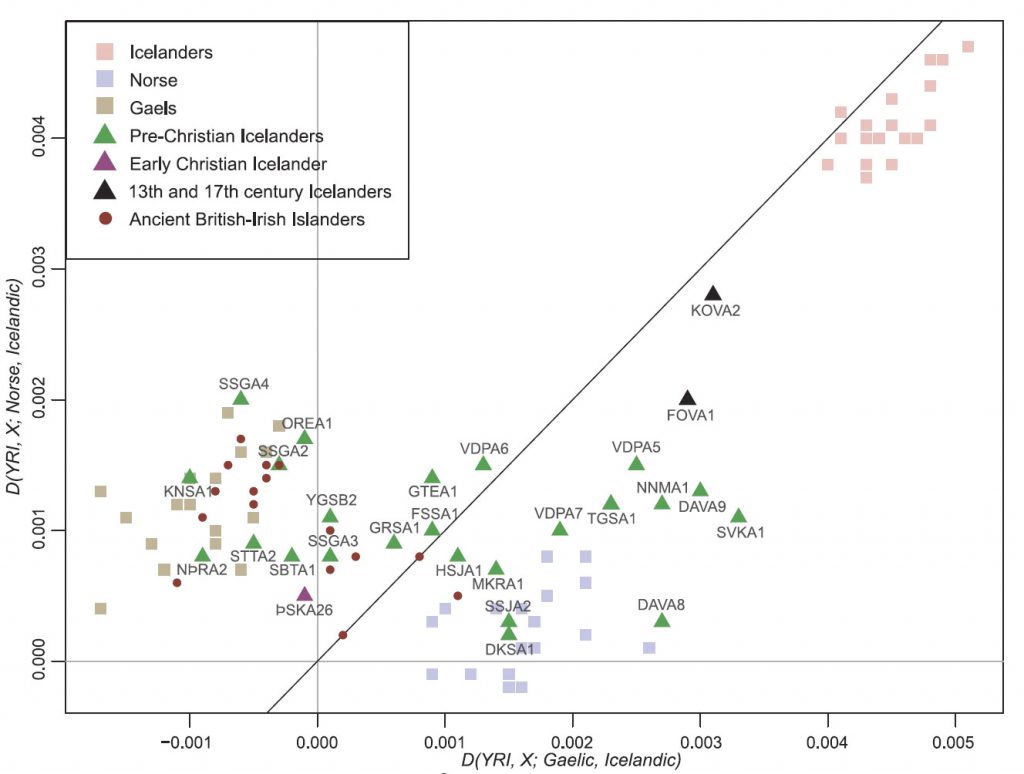
We estimated the mean Norse ancestry of the settlement population (24 pre-Christians and one early Christian) as 0.566 [95% confidence interval (CI) 0.431–0.702], with a nonsignificant difference betweenmales (0.579) and females (0.521). Applying the same ADMIXTURE analysis to each of the 916 contemporary Icelanders, we obtained a mean Norse ancestry of 0.704 (95% CI 0.699–0.709). Although not statistically significant (t test p = 0.058), this difference is suggestive. A similar difference ofNorse ancestry was observed with a frequency-based weighted least-squares admixture estimator (16), 0.625 [Mean squared error (MSE) = 0.083] versus 0.74 (MSE = 0.0037). Finally, the D-statistic test D(YRI, X; Gaelic, Norse) also revealed a greater affinity between Norse and contemporary Icelanders (0.0004, 95% CI 0.00008–0.00072) than between Norse and ancient Icelanders (−0.0002, 95% CI −0.00056–0.00015). This observation raises the possibility that reproductive success among the earliest Icelanders was stratified by ancestry, as genetic drift alone is unlikely to systematically alter ancestry at thousands of independent loci (fig. S10). We note that many settlers of Gaelic ancestry came to Iceland as slaves, whose survival and freedom to reproduce is likely to have been constrained (17). Some shift in ancestry must also be due to later immigration from Denmark, which maintained colonial control over Iceland from 1380 to 1944 (for example, in 1930 there were 745 Danes out of a total population of 108,629 in Iceland) (18).
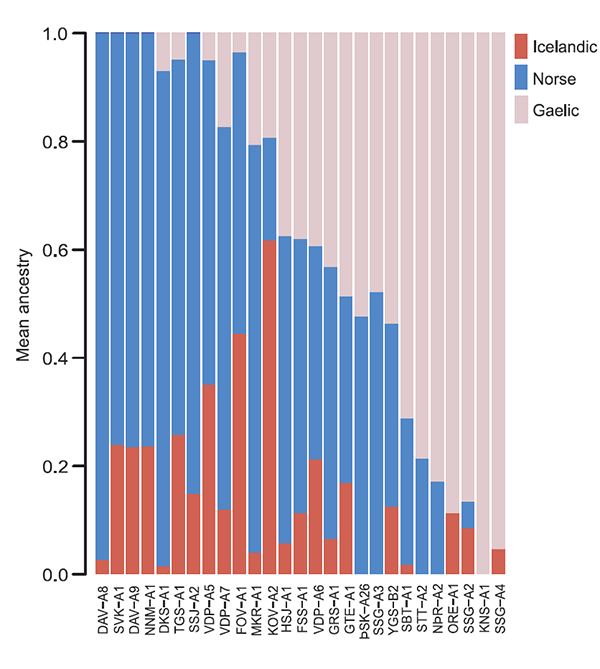
Gaelic, and Icelandic ancestry for ancient Icelanders using ADMIXTURE
in supervised mode.
Five pre-Christian Icelanders (VDP-A5, DAVA9, NNM-A1, SVK-A1 and TGS-A1) fall just outside the space occupied by contemporary Norse in Fig. 3A. That these individuals show a stronger signal of drift shared with contemporary Icelanders is also apparent in the results of ADMIXTURE, run in supervised mode with three contemporary reference populations (Norse, Gaelic, and Icelandic) (Fig. 3B). The correlation between the proportion of Icelandic ancestry from this analysis and PC1 in Fig. 2A is |r| = 0.913.(…)
(…) as the five ancient Icelanders fall well within the cluster of contemporary Scandinavians (Fig. 3C), we conclude that they, or close relatives, likely contributed more to the contemporary Icelandic gene pool than the other pre-Christians. We note that this observation is consistent with the inference that settlers of Norse ancestry had greater reproductive success than those of Gaelic ancestry.
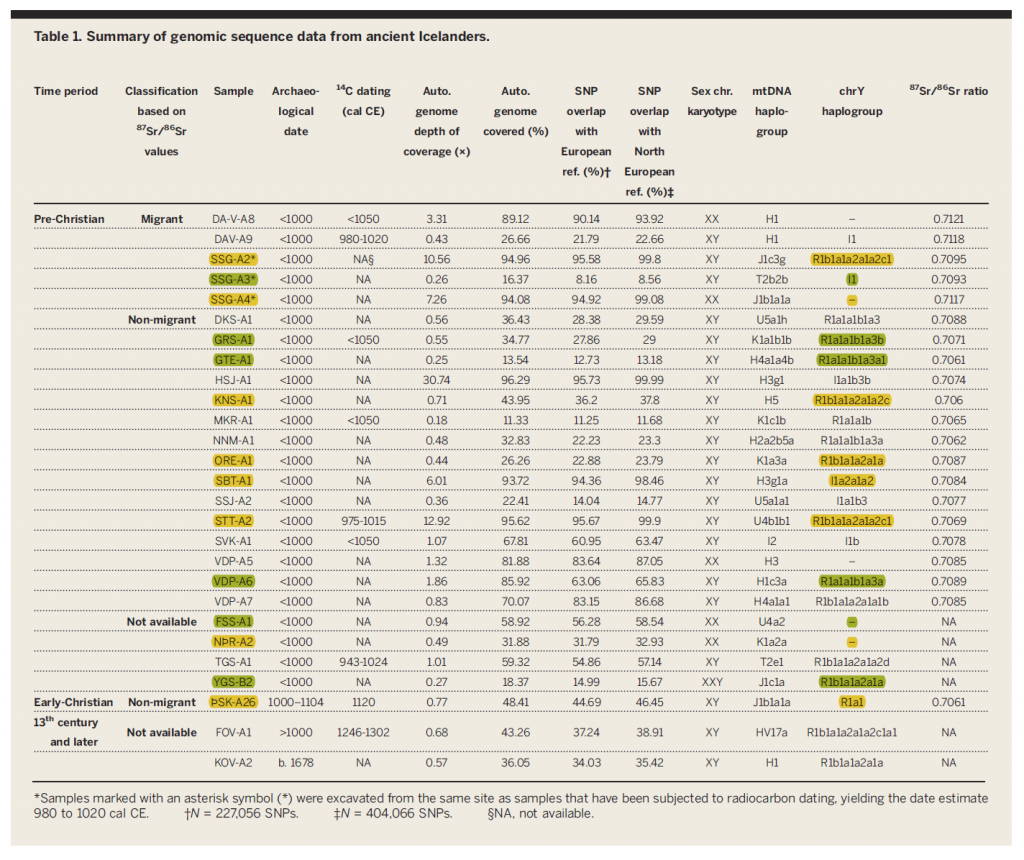
Ancient Icelanders show a clear relation with the typically Norse Y-DNA distribution: I1 / R1a-Z284 / R1b-U106.
- Among R1a, the picture is uniformly of R1a-Z284 (at least five of the seven reported).
- There are six samples of I1, with great variation in subclades.
- Among R1b-L51 subclades (ten samples), there are U106 (at least one sample), L21 (three samples), and another P312 (L238); see above the relationship with those clustering closely with Gaelic samples, marked in fluorescent, which is compatible with Gaelic settlers (predominantly of R1b-L21 lineages) coming to Iceland as slaves.
Probably not much of a surprise, coming from Norse speakers, but they are another relevant reference for comparison with samples of East Germanic tribes, when they appear.
Also, the first reported Klinefelter (XXY) in ancient DNA (sample ID is YGS-B2).
Related:
- Genomic analysis of Germanic tribes from Bavaria show North-Central European ancestry
- Germanic tribes during the Barbarian migrations show mainly R1b, also I lineages
- mtDNA suggest original East Germanic population linked to Jutland Iron Age and Bell Beaker
- Admixture of Srubna and Huns in Hungarian conquerors
- The concept of “Outlier” in Human Ancestry (III): Late Neolithic samples from the Baltic region and origins of the Corded Ware culture
- Genetic prehistory of the Baltic Sea region and Y-DNA: Corded Ware and R1a-Z645, Bronze Age and N1c
- Bell Beaker/early Late Neolithic (NOT Corded Ware/Battle Axe) identified as forming the Pre-Germanic community in Scandinavia
- The Tollense Valley battlefield: the North European ‘Trojan war’ that hints to western Balto-Slavic origins